Alpine Tundra Soil at Spring Snow Melt Harbors Novel and Abundant Groups
of Chloroflexi Bacteria
Poster N-252 ASM General Meeting 2003
Elizabeth K. Costello
and Steven K. Schmidt
Department of Ecology and Evolutionary Biology, University of Colorado, Boulder
Introduction
Tundra soil ecosystems play critical roles in atmospheric and biogeochemical
nutrient cycling and are particularly sensitive to impacts of pollution and
climate change. Despite extensive research on alpine tundra biogeochemistry
at the Niwot Ridge Long-Term Ecological Research site, we know almost nothing
about the phylogenetic diversity and community structure of the microorganisms
mediating these biogeochemical processes.
In addition, alpine tundra is an unexplored microbial habitat, which may harbor
unique microorganisms. The discovery of novel lineages has inherent value to
our understanding of biodiversity. We examined bacterial diversity and community
structure across a depth profile in an alpine tundra wet meadow soil at the
time of snowmelt.
- Wet meadows occur down slope from snow beds and receive cold water drainage
throughout much of the growing season such that soils remain near saturation
(2,6).
- Plant productivity, soil organic matter, and microbial activity are highest
in wet meadows compared to drier alpine tundra communities (2).
- Previous studies on tundra soils at our site have shown a significant microbial
community shift and the appearance of novel bacteria associated with snowmelt
(5).
Objectives
- Make a general survey of bacterial diversity using culture-independent
16S rDNA based techniques.
- Identify novel groups via phylogenetic analysis and target these groups
for further investigation.
- Examine bacterial community variation across depth (and eventually season)
using t-RFLP analysis.
- Correlate the diversity and distribution of a novel group with soil characteristics.
Methods and Results
Objective 1. Make a general survey of bacterial diversity
Bacterial 16S rDNA library construction:
- A 20 cm soil core was collected in early June, when water was flowing over
the saturated soil. DNA was extracted from soil core segments at ~ 2 cm intervals.
- A composite DNA sample was used as template for amplification of bacterial
16S rDNA using 8F (5’) and 1391R (3’) primers. 137 ~1300 bp sequences
were aligned in ARB and analyzed using Bayesian phylogenetic inference. RDP
(8) and Bellerophon (7) were used for chimera checking.
Results:
- Fifteen “division level” clades were represented. Divisions
occurring most frequently were the Acidobacteria (26%), Proteobacteria
(24%), Chloroflexi (16%), Cytophaga-Flexibacter-Bacteroides
(9%), and the Verrucomicrobia (7%) (Figure 1).
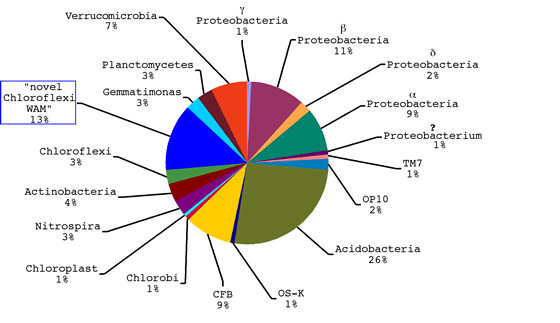
Figure 1: “Division” level representation of bacterial 16S rDNA
sequences from alpine tundra wet meadow soil at spring snow melt. Sequences
were aligned against an ARB database. Percentages were derived from Bayesian
phylogenetic inference of 244 total sequences, including representatives from
34 bacterial divisions, for 6.0 x 106 generations (tree available). Blue box
indicates novel group WAM (wet alpine meadow).
- Eighteen novel clones (13%) with strong support for an affiliation with
the Chloroflexi were also discovered. Two distinct clades were evident
and are referred to as “WAM1” and “WAM2” (wet alpine
meadow clones).
Objective 2. Investigate diversity and distribution of novel groups WAM1 and
WAM2
Specific amplification and phylogenetic analysis sequences from a novel group:
WAM1 and WAM2 5’ 16S rDNA primers were designed and paired with 1391R
(3’).
- Hotstart/touchdown PCR was used to specifically amplify 16S rDNA from the
novel groups using DNA templates from each soil depth sample and samples from
fall and winter.
- WAM1 and WAM2 sequences were aligned in ARB and trees were generated using
Bayesian phylogenetic inference.
Results:
- Primers targeted to the WAM1 and WAM2 clades specifically amplify the correct
target sequence and expand the diversity of the groups. 54 WAM1 sequences
and 30 WAM2 sequences all have the correct primer site (Figure 2).
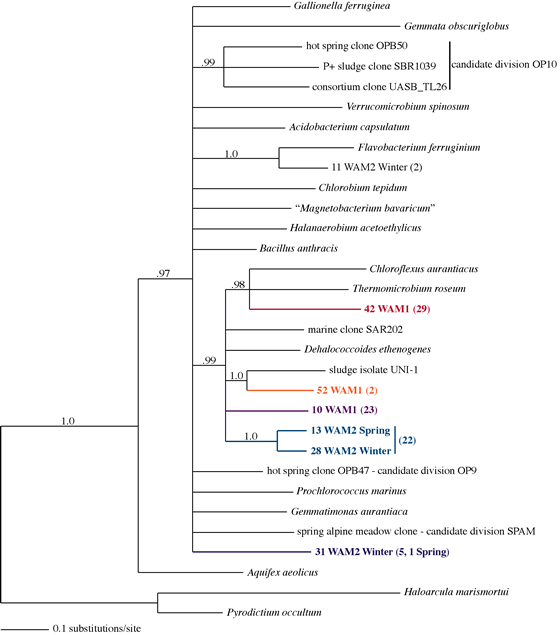
Figure 2: Bayesian tree of ARB aligned WAM1 and WAM2 sequences generated using
specific primers and several representative sequences. Parentheses indicate
number of clones analyzed from each clade. WAM2 sequences were found in soil
collected at spring snow melt and in frozen surface soil collected in the winter
under snow. Nodes with posterior probabilities >= .95 are shown.
- Six WAM2 clones appear to form a new candidate division of Bacteria (Figure
2).
- WAM1 and WAM2 generate amplification product in PCR with subsurface DNA
and not from surface samples in the spring core (Figure 3). WAM1 product appears
in spring and not fall; WAM2 product appears in winter but not fall (Figure
3).
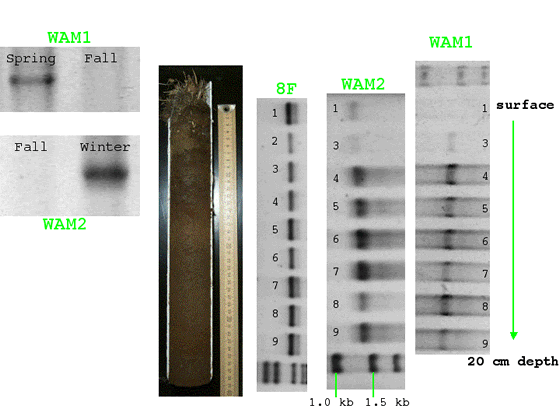
Figure 3: Electrophoresed PCR product from specific primers WAM1, WAM2, and
universal bacterial primer 8F PCR. Photograph of soil core at center.
Objective 3. Examine bacterial community structure across depth
Terminal Restriction Fragment Length Polymorphism (t-RFLP) Analysis:
- Labeled *8F primer was used with 1391R to generate labeled PCR product
for each soil core segment.
- Hae III was used to digest the labeled PCR products. Terminal
restriction fragment profiles were separated on a polyacrylamide gel and compiled
using LiCor GeneImagIR software.
- Fragment lengths were coded as characters in MacClade and PAUP parsimony
analysis was used to infer relatedness among community profiles across depth
at 2 cm intervals.
Result:
- Soil depth profile t-RFLP analysis of bacterial 16S rRNA genes confirms
a substantial shift in community composition at 1-5 cm, below which the t-RFLP
pattern remained more similar at 2 cm intervals to 20 cm (Figure 4).
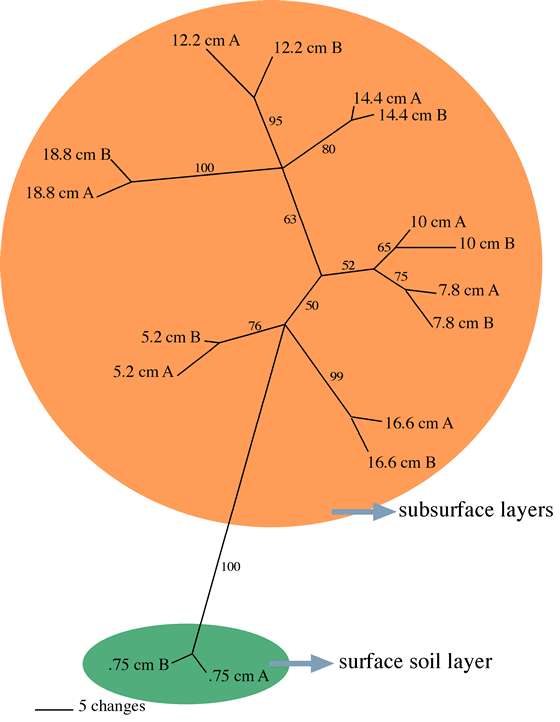
Figure 4: Parsimony tree demonstrating relationships between t-RFLP profiles
for segments of a soil core collected at spring snow melt. Bootstrap values
>= 50 are shown (10,000 replicates) and nodes with less that 50% bootstrap
support are collapsed.
Objective 4. Correlate the diversity and distribution of a novel group with
soil characteristics.
Preliminary measurements of soil organic matter (SOM), moisture, Fe, Mn, and
SO42+ concentrations were made using standard techniques.
Results:
- SOM and soil moisture are highest at the surface (Figure 5a).
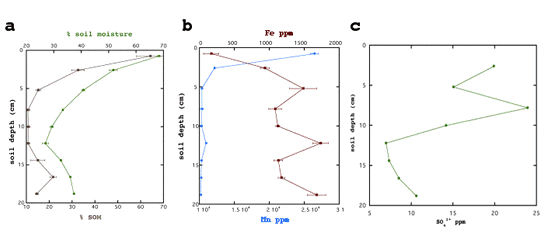
Figure 5: Spring soil core segment organic matter, moisture, Fe, Mn, SO42+ are
plotted against segment depth (left axes).
- Fe increases with depth while Mn decreases (Figure 5b).
- SO42+ generally decreases with depth but shows an increase around 8 cm
(Figure 5c).
Discussion
- Our study reveals previously unknown groups of bacteria (WAM) in a single
alpine tundra soil core sampled at spring snow melt, highlighting the continued
importance of general culture-independent 16S rDNA surveys of unexplored environments.
- These novel bacteria make up a substantial proportion of the diversity sampled
and are likely to play key roles in alpine tundra soil biogeochemistry.
- The known diversity of Chloroflexi bacteria (see 1, 3, 4) is expanded by
this study. WAM sequences largely represent new groups of Chloroflexi.
- We show that the abundance of WAM is structured by depth in the spring.
WAM also appears in surface soil under snow in winter, but seems to decrease
in abundance in fall after the summer growing season. WAM may dominate anaerobic
wet meadow soil. WAM primers have also amplified sequences from Peru and Antarctica
(sediments, data not shown). Therefore, WAM may be globally abundant.
- Our t-RFLP data also shows a major community shift between surface and subsurface,
most likely due to a redox gradient in this soil.
- Finally, WAM is correlated with lower soil organic matter and moisture and
higher Fe content. WAM may use Fe or other metals for anaerobic respiration.
Future work
We hope to investigate the distribution, morphology, physiology, and population
dynamics of WAM further via correlations with soil characteristics, FISH with
confocal imaging, culturing, and genomics techniques.
Works cited:
1. Björnsson, L. et al. 2002. Microbiology. 148(8):2309-18
2. Fisk, M.C. et al. 1998. Ecology. 79(7):2253-66
3. Giovannoni, S.J. et al. 1996. Proc. Natl. Acad. Sci. USA. 93:7979-84
4. Hugenholtz, P. et al. 1998. Journal of Bacteriology. 180(18):4765-74
5. Lipson, D.A. et al. 2002. Microbial Ecology. 43:307-14
6. West, A.E. et al. 1999. Biogeochemistry. 45:243-64
7. http://foo.maths.uq.edu.au/~huber/bellerophon.pl
8. http://rdp.cme.msu.edu/html/index.html
 This research
was supported by an NSF Microbial Observatories Grant.
This research
was supported by an NSF Microbial Observatories Grant.





 This research
was supported by an NSF Microbial Observatories Grant.
This research
was supported by an NSF Microbial Observatories Grant.