I am a PhD student in Dr Thomas Huber’s Biomolecular Modelling Group within the Research School of Chemistry (RSC) at the Australian National University (ANU). The members of my associate supervisory team are Dr Nicholas Hamilton and Dr Justine Hill. I collaborate closely with Professor Gottfried Otting and his Group at ANU. My PhD thesis is titled: “Computing protein interactions using NMR effects from paramagnetic labels”.
Paramagnetic effects provide rich structural information in the form of long-range distance and, or, orientational information not available from conventional Nuclear Magnetic Resonance (NMR) measurements. When a paramagnetic metal ion, such characterised by unpaired electrons, is introduced into a protein, facilitated through the availability of a native binding site, or by exploiting readily available tagging agents, the pseudocontact shift (PCS), paramagnetic relaxation enhancement (PRE) and residual dipolar coupling (RDC) are features readily measured in NMR experiments . These effects are becoming frequently exploited in problems such as the assignment of spectra, the characterisation of molecular interactions, and in the detection and reconstruction of protein dynamics.
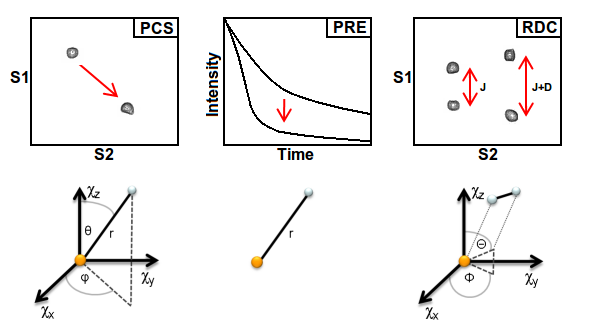
Figure 1. The changes in a NMR spectrum as the result of of a paramagnetic ion. Schematic illustration of the relationship between the position and, or, orientation of the paramagnetic ion (orange sphere) and nuclear active spin(s) (white sphere) probed by the new experimental observables.
The problems I am investigating are: