Bellerophon
Detection of chimeric sequences
What are Bellerophon webservices?
The Bellerophon web service allows remote object execution of Bellerophon modules in order to integrate chimera detection in complex high-throughput 16S rRNA processing workflows.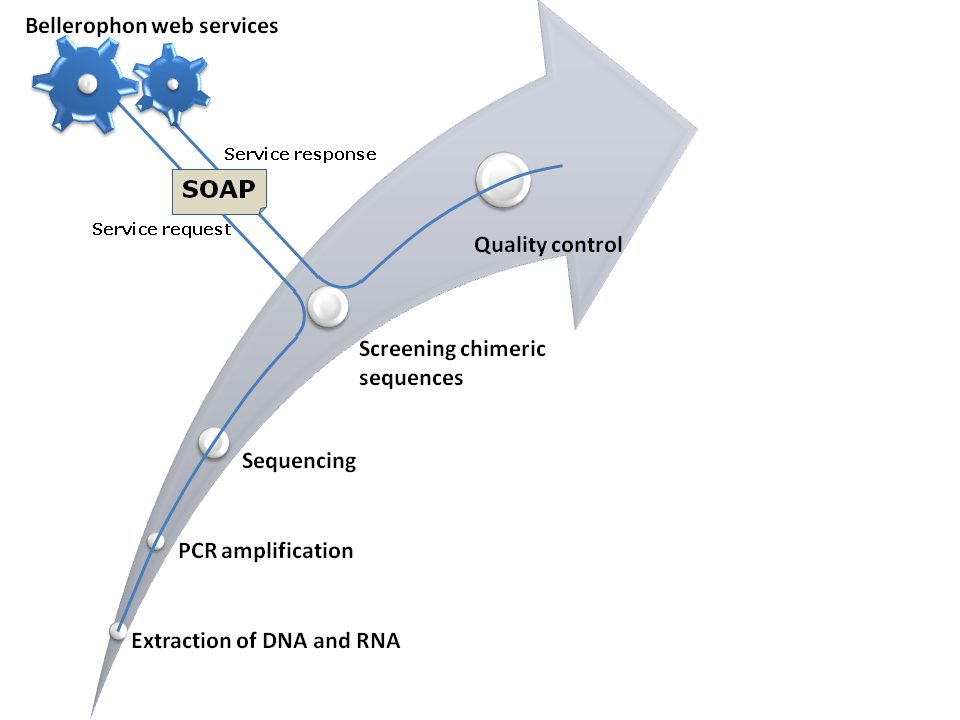
More information on Bellerophon is available on the Bellerophon website and a excellent introduction to web services is provided by the European Bioinformatics Institute.
What software is required?
We highly recommend to use Python and Zolera SOAP Infrastructure (ZSI) to communicate with Bellerophon web services, and we provide example python scripts using ZSI to demonstrate how simple it is to implement Bellerophon requests in your application.We also provide a Web Services Description Language (WSDL) interface description file to implement service calls with any other programming language.
Do I need to register?
For funding purpose we like to keep track of who uses our Bellerophon service. A simple key system is used to identify you as a user and you can obtain a key free of any charge here.How to get started?
To get started quickly, we provide example python scripts using ZSI. To run these examples, the following steps are required:1) install python and ZSI, if they are not already installed on your computer
2) download the python script client_Bellerophon.py and the WSDL file Bellerophon.wsdl
3) generate python SOAP interface files with 'wsdl2py -b Bellerophon.wsdl'
4) edit the script file client_Bellerophon.py to include your key where indicated in the script.
The Bellerophon key can be obtained here
5) run the client script (type 'python client_Bellerophon.py' on the command line). An example input FASTA file can be found here.
after a few minutes you should receive the results.
Please be considerate
Bellerophon calculations (in particular for sequences searched not within a PCR library) are computationally intensive and require significant CPU time. We allow PCR libraries up to 1000 unaligned sequences (1500 aligned sequences), or up to 100 non-PCR library sequences to be processed per submission. In non-PCR library searches, Bellerophon3 is used to evaluate each sequence against the greengene database (currently > 400000 sequences), which typically takes approximately one minute per sequence. Please be considerate with submitting many sequences, because our resources are limited and your submission will also effect other users.How to read the results?
Please read the Bellerophon documentation for more details on the output and the original Bellerophon publication on algorithmic details.Reference to cite:
T. Huber, G. Faulkner and P. Hugenholtz. Bellerophon; a program to detect chimeric sequences in multiple sequence alignments, Bioinformatics (2004) 20, 2317-2319.T. Huber, T.Z DeSantis and P. Hugenholtz. Bellerophon web services; integration of chimeric sequence detection in high-throughput r16S gene analysis pipelines, Nucl. Acid Res. (2011) Webserver issue, to be submitted.